How to Use the Autosomal DNA Segment Analyzer
I created this tool in an attempt to put all the relevant information available that was needed to evaluate segment matches on a single, interactive web page. It relies on the three files for a single test kit that DNAgedcom.com collects from FamilyTreeDNA.com. These files include information about your matches, matching segment locations and sizes, and "in common with" (ICW) data. Using these files, the tool will construct a table for each chromosome which includes match and segment information as well as a visual graph of overlapping segments, juxtaposed with a customized, color-coded ICW matrix that will permit you to triangulate matching segments without having to look in multiple spreadsheets or on different screens in FamilyTreeDNA. Additional information, such as ancestral surnames, suggested relationship ranges, and matching segments and ICWs on other chromosomes is provided by hovering over match names or segments on the screen. Emails to persons you match may also be generated from the page. The web page produced by this program does not depend on any other files and may be saved as a stand-alone .html (or .htm) file that will function locally (or offline) in your browser. You can even email it to your matches as an attachment. You can play with a working sample output here.
To access the tool, first download the three FamilyTreeDNA files for your kit at www.DNAgedcom.com:
| [kitnumber]_icw.csv | a spreadsheet containing "In Common With" matches |
| [kitnumber]_ChromosomeBrowser.csv | a spreadsheet that lists all matching segments on every chromosome pair |
| [kitnumber]_Family_Finder_Matches.csv | a spreadsheet listing all matches for the test kit |
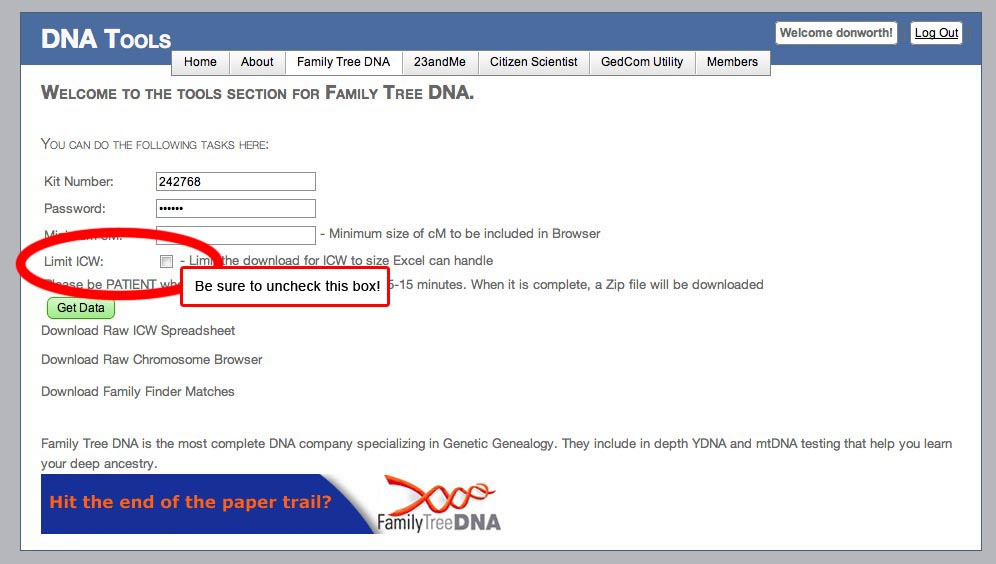
When using DNAgedcom, create a free account there if you haven't already and logon. Then select the "Family Tree DNA" tab and pick "Download Family Tree DNA Data". Set up your input screen like this:

Fill in your own FamilyTreeDNA kit number and password as I did above. It may prompt the DNAgedcom userid - if so, erase that and replace it with your kit number instead. Do not prefix it with an "F". You can put a 5 into the Minimum cM box if you wish, but it might be better to have all the data for other future uses, so I recommend leaving that box blank. BE SURE TO UNCHECK THE LIMIT ICW BOX! Otherwise your ICW matrices may be only partially filled in. When DNAgedcom finishes, it will download a single, compressed ZIP archive file to your computer. Unzip it to get your three files and put them in a safe place (I usually make a folder for them labeled with the name of the person I tested, kit number, and date). If you have any problems getting the files, consult the "IF THINGS DON'T WORK FOR YOU" box at the bottom of this page.
| Kudos where kudos are due! There would be no ADSA tool if it were not for DNAgedcom. Rob Warthen of DNAgedcom has done us all a wonderful service by gathering the match, ICW and segment data from FamilyTreeDNA. Please support DNAgedcom by using the DONATE button on its home page! |
Once you have the three files, direct your browser to:
Assuming you are not logged on in DNAgedcom at the time, you will see a screen like this (if you are logged on, you will see a slightly different screen, but I will cover that when I talk about the input files below):
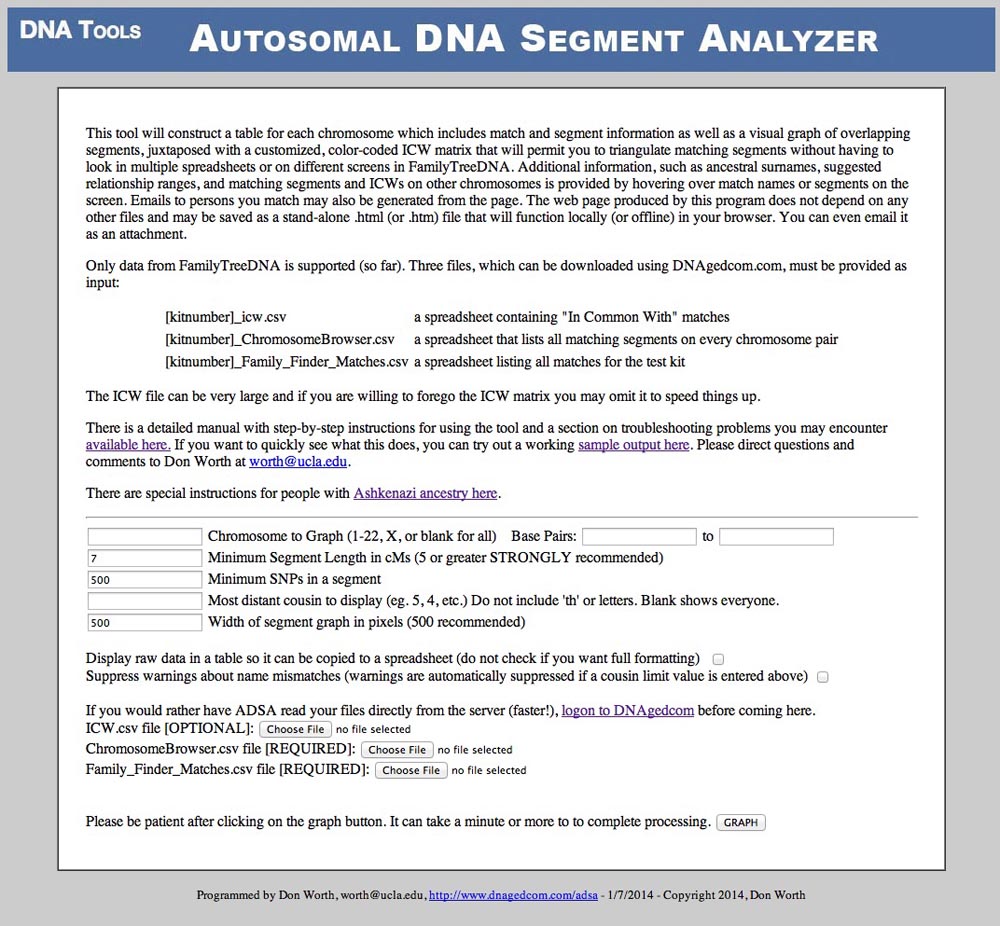
The inputs on this screen are as follows. Other than specifying the names of the files you want to use, you can just accept the default value for the rest of these:
Chromosome to Graph
You may specify a single chromosome to graph (valid inputs are 1 through 22 or X) or leave this box blank to see segments for all 22 autosomal chromosome pairs and the X chromosome. The default prompt is blank - generate tables for every chromosome. If you specify a single chromosome to graph, you may also "zoom in" further by giving a starting and ending base pair position. The tool will ignore any segments that do not at least partially overlap that region on the chromosome. This is a way to reduce the size of the output file if you have so many segments that the browser bogs down looking at them (if you set minimum segment length to 1 cM, for example, or if you have Ashkenazi ancestry). If you leave the start box blank, 0 is assumed. If you leave the end box blank, 300,000,000 is assumed.
Minimum Segment Length in cMs
The tool will ignore any sgements in the Chromosome Browser file that are smaller than the number you specify here. You should not go below 5 cMs because the resulting page becomes so gigantic that your browser will be almost too sluggish to use. The prompt is 7 cMs.
Minimum SNPs in a segment
The tool will ignore any segments with fewer than the number of SNPs you specify here. The prompt is 500.
Most Distant Cousin to Display
This could be useful to anyone at one time or another, but especially to those with gigantic files (such as persons with Ashkenazi ancestry). You can ask ADSA to ignore matches that are distantly related by entering a "cousin number" here. Entering 1 would be 1st cousin, 2 would be 2nd cousin, and so on. Blank is everyone. Zero is anyone more closely related than 1st cousin (sibling, parent, aunt or uncle, etc.) So if you enter a 3, then all matches with a Family Tree DNA predicted relationship of 3rd cousin or closer will be included. Predicted 4th, 5th and distant cousins will be skipped. (Side note: doing this makes it impossible for the program to check to make sure there are no name mismatches between the files, so, to prevent a torrent of warnings about that, the warning suppression option is turned on regardless of what you put in its checkbox.) If you have huge files and ADSA has been timing out or giving 500 Internal Server Error messages, try starting with 2nd cousin on this input and if it works, you can "walk" out further (3, 4, etc.) until you know what the highest value that works will be.
Width of Segment Graph in Pixels
The wider you go, the easier it is to see the segments spread out across the length of the chromosome. However, if you go too wide, it may be difficult to print the page without having the browser chop things off at the right margin. The default here (500 pixels) seems to work pretty well in most cases for me (although I might have to print in landscape when I have a lot of segments on one chromosome). Once in a while you may want to make the segment graph smaller if you have a chromosome with over 50 segments on it since the ICW matrix will tend to push the table out beyond the right margin for printing.
Display raw data in a table so it can be copied to a spreadsheet
The ICW matrix is generated with web graphics and cannot be copied to the clipboard. Also, some of the fixed width formatting of the text table to its left causes problems with copy and pasting data into a spreadsheet. If you want to capture the segment data and ICW matrix for a spreadsheet, check this box. But be warned, you will just get a big table full of text and numbers - no hovering, graphing or colors. (Note: blank/empty names and email addresses will be replaced with a question mark to preserve the columns in Excel.) The default is to leave this box unchecked so that the fully formatted, colored tables and graphs are generated.
Suppress warnings about name mismatches
Sometimes ADSA is unable to relate a row in the ICW or Chromosome Browser files back to a row in the match table because the name of the person tested is spelled differently in the various files. If that occurs, ADSA will issue a warning message on the output page and ignore the row with the problem. If you have already seen the warnings and are planning to save or print the output page, then you may not want the warnings to be displayed. If so, check this box.
With regard to the input files, you have two options. If you are not logged into DNAgedcom when you arrive on the ADSA input form page, you will see prompts to show ADSA where the three individual files are on your computer's hard drive. (That's the version of the screen above). In that case, once you have browsed to each file ADSA will be ready to upload them to the server and run the program against them. However, there is another option. When you first downloaded the files from DNAgedcom, in addition to downloading the zip file, the server also stored all three files for you, associated with your DNAgedcom logon ID. Instead of uploading them from your computer, you may, instead, point ADSA at those server-side (techie lingo!) copies of your files and the upload step can be skipped entirely. This might be a better option if you have very large files. Even if you don't, running ADSA will probably be about ten times faster if it doesn't have to upload your files back to the very same server that produced them in the first place! In fact, if you have Ashkenazi ancestry, skipping the upload like this may be the only way you can even get ADSA to work on your files. Ashkenazi files can get so huge that the server will time-out in the process of attempting to upload them. The main reason for the upload option is to allow people to edit their files before feeding them into ADSA. (Although you also have the option to load them back onto the DNAgedcom server after you edit them on the VIEW FILES panel.)
If you want to try the faster way, go to www.DNAgedcom.com and logon with your logon ID and password. Before you go to ADSA, navigate to the MEMBERS menu (upper right corner of the screen) and select VIEW FILES. You should see a folder in the left panel that is named something like FTDNA_242768 (except the numbers will be your kit number, not mine). If you click on that folder you should see your three files, stored there already. (If you see them in a folder called "building" that means that one or more of the files may be only partially written - DNAgedcom didn't finish downloading them from FamilyTreeDNA - so try that step again). Assuming you find the three files, you can point your browser at ADSA - www.DNAgedcom.com/adsa - and you should see this at the bottom of your input form, replacing the prompts for the three files:

All you nave to do here is type in your FamilyTreeDNA kit number. ADSA will find all three of the files it needs on the server from that.
NOTE: If you downloaded your files prior to 1/13/2014 your ICW file may be MUCH larger than either of the other two files (my wife's is 4.6 MB because she has kits for two sisters and an aunt that greatly increased the number of rows in her ICW file), and it can take a minute or two - or more sometimes - for it to upload to the server. People with Ashkenazi ancestry can have ICW files of upwards of 300 MB or more and ADSA will timeout with a blank screen or 500 Internal Server Error if you attempt to upload files that large. It is strongly recommended that you re-download your files now because you will find that the new ICW file is more than 10 times smaller than before that date due to some enhancements that were added to the DNAgedcom download utility then. If you're in a hurry, you can skip the ICW file (on the version of the ADSA screen that uploads the files), but you won't get the nice ICW matrixes if you do! Both the other two files (the Chromosome Browser and Matches files) are required for the tool to work. There is no option to skip the ICW file if you are logged on to DNAgedcom when you get to ADSA.
When you're ready, click on the GRAPH button and the tool will read your files and generate the web page. It may be just a few seconds, or it may take a minute or more if your ICW file is several megabytes in size and/or you have a lot of segments. If you run into trouble at this point or you see a lot of warnings, consult the "IF THINGS DON'T WORK FOR YOU" box at the bottom of this page.
This is a sample of what you'll see (I have blurred out the names to protect the innocent):
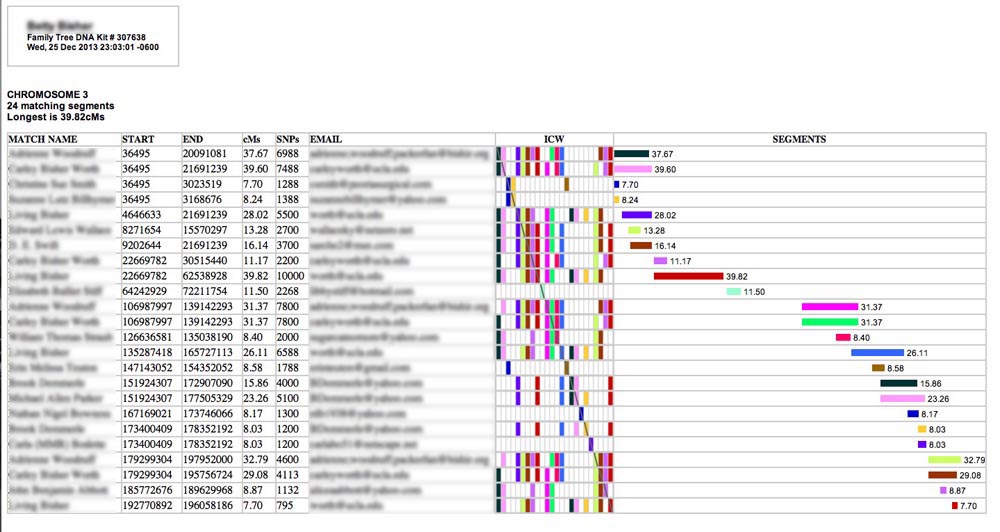
Each row represents a separate matching segment on the chromosome. The textual information in the left-hand columns describes the segment. The match person's name and email are also provided. (If a name or an email address is too long, it will be truncated - an ellipses indicates this - but you can see the full entry by hovering your cursor over it for a moment.) In the center of the table is an ICW matrix (more about that later), and on the right is a graphical representation of the segments annotated with their length in cMs.
Hovering your cursor over one of the names on the left will produce a pop-up "tool tip" something like this:

In addition to the match's name and email address the pop-up includes the date the match occurred, the predicted relationship range, and the list of ancestral surnames. (Sorry, the GED file is not included in the files DNAgedcom produces!)
Hovering your cursor over one of the colored horizontal bars on the right side under "SEGMENTS" will produce a pop-up like this one with even more information:
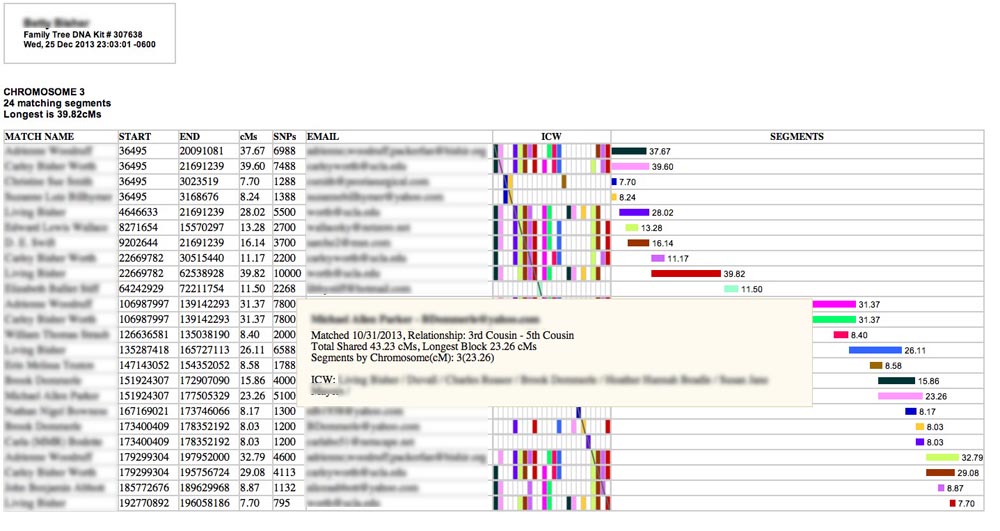
Here you will see Total Shared cMs for this person (across all the chromosomes), the longest block you share with them in cMs (which may or may not be on this chromosome), and a list of all the matching segments you have with this person. (In this case I had only one shared segment - and it is on this chromosome.) Also listed are ALL the ICW names for this person (even if they are due to matching segments on other chromosomes).
If you click on one of the emails in the email column of the table, you will be transferred to your email program with the TO: line containing the match name and the email address. (FTDNA does this also, but does not include a name with the email address, which, I have found, can sometimes cause an email to end up in the recipient's SPAM folder!)
The segments are plotted on the right of the ICW matrix, scaled to base pairs (so, as with the FTDNA Chromosome Browser, the apparent length of the segment may or may not be representative of the actual cMs) and, to save on horizontal screen space, I did not include any empty portions of the chromosome prior to the first segment or after the last one. If there are only a few matching segments on a chromosome, this can sometimes make relatively short segments look very long, since they expand to fill the width of the table. However, the main purpose of the graph is to visualize overlaps and I was trying to avoid clipping at the right margin during printing.
The ICW matrix has been generated to include ICW information only for the persons associated with the matching segments listed for this chromosome. The colors are intended to make triangulation easier. Look at the example below.

(I have added the colored shading and arrows to illustrate the process - they do not appear on the ADSA output). All of the segments on the far right of the figure above appear to overlap one another - they are "stacked" and generally occupy the same locations along the chromosome. Let's start with Bugs Bunny's segment (the yellow-green one with a length of 16.75 cMs). If you follow Bugs' row to the left from his segment into the ICW matrix you will eventually arrive at the cell in the matrix where Bugs' row meets his column. This cell is marked with a backwards slash. If you then travel either up or down Bugs' column you will see that the color of the little "bricks" in his ICW column matches that of Bugs' horizontal segment on the far right (yellow-green) forming a sideways "T". Each segment that is opposite a yellow-green "brick" in Bugs' column is ICW to Bugs. That is to say, those segments belong to people who match Bugs and match you (the person tested) - they are matches you have in common with Bugs. For example, you'll notice that there are yellow-green bricks opposite the rows of Fred Flintstone, Betty Rubble, Daffey Duck, and Huckleberry Hound. All four of them are matches to you and are matches to Bugs Bunny. You, Bugs and Fred are said to be "triangulated" since there are three of you and you all match each other. The same is true for you, Bugs and Betty or Daffey, or Huckleberry. Assuming the segments are long enough to be IBD (and they appear to be) and that none of you is double-related, that means that you, Bugs and Fred all share a common ancestry. In fact, you can see that the ICW columns for Betty Rubble (dark purple), Bugs (yellow-green), Daffey Duck (brown), and Huckleberry Hound (magenta) form a multi-colored rectangle, or block of ICW "bricks". That rectangular collection of ICW "bricks" means that you all ICW each other and all of you probably share the same common ancestry. Look above Betty and you will see that Fred Flintstone also has the same configuration of colored bricks along his row as the others - he is part of the group as well. I have shaded all these ICWed rows pink. Now look at Wilma and Barney. While their segments look like they completely overlap those of the others, they do not participate in the ICW block for the pink group. In fact, they form their own little 2x2 block of bricks, meaning they match you and each other, but they do not match Fred, Betty, Bugs, Daffey or Huckleberry. Because they occupy the same place on the chromosome as the others, its clear that they are matching the other chromosome of the pair. If the pink group matches, say, the chromosome you got from your mother, then the blue group matches the one you got from your father (or vice versa).
When you are looking at ICW "bricks" you'll notice that almost all of the time, the patterns are symmetrical around the diagonal line. If A matches B, then B matches A. So if you see a colored brick - let's say the pink one to the left of Bugs Bunny's "slash", you can determine whose brick that is by following that column up or down until you hit a pink brick with a slash through it. In this case, that happens on Fred Flintstone's row - so Fred matches Bugs. And, on Fred's row, you will see that there is a light green brick in Bugs' column. The mirror image of the pink one we started with. The symmetry is a little hard to visualize because the bricks aren't square. (Rarely, there is a brick that doesn't have a symmetrical "partner". This seems to be due to a glitch in the data coming from FamilyTreeDNA. My guess is that the matches for the two kits were computed at different times with slightly different match criterea.)
TIP: If you have trouble seeing the ICW cells, you can use your browser's enlarge feature (ctrl + on Windows or Command + on MacOS) to zoom in on it. I also use ctrl - (or Command - on the Mac) to shrink the screen for chromosomes with a lot of segments to see all of them on the screen at once.
Often you will see patterns in the ICW matrix like this:

Look for small rectangles like the one in the upper left that is outlined in red. That 4x4 block represents four segments that all match each other and match your kit. Occasionally you will get something like the big pattern in the upper right. These big blocks of bricks are often associated with fairly short segments, so be sure the segment lengths are long enough to be IBD. (IBS segments often stack up like that - try running with minimum cM at 2 or 3 and you will see a lot of them!). Remember that 50% of the time 8 cM segments are IBS and you don't get very close to 100% certainty of IBD with segments smaller than 10 cM. However, if this configuration is truely IBD, my guess is that the common ancestor has a LOT of descendants in the general population! I figure the more matches I can find on one segment, the more help I will have from those people in finding the common ancestor. The one on the bottom is the pattern to watch out for. Clearly that brown 8.27 cM segment outlined in red that doesn't match the others is matching on the chromosome of the pair that came from my other parent.
Here is another case to be aware of. Suppose you match Fred Flintstone and Barney Rubble and they are ICW to each other. But when you go look for Fred, the common segment he shares with you is on chromosome 11 but you don't see Barney on that chromosome at all. And when you look for Barney his segment is on chromosome 21, but Fred isn't there at all. You know Fred matches Barney because the ICW tells you so. How come you don't see a segment that they both match? It's important to remember that ICWs tell you that two of your matches match each other - but they do not tell you how or where they match. Often two ICWs will match each other on the same segment they share with you. But sometimes they match on another segment (or chromosome) entirely - one that you, yourself don't have and that you therefore cannot see. In fact, they may be matching each other "right under your nose" in the sense that, for example, you got the right side of a longer segment which you share with Fred, and Barney got the left half of that segment and shares that (invisibly to you) with Fred. So Fred may have the entire length of that segment. You were just lucky that you matched Barney on some other segment in your output or you wouldn't even know about his match to Fred. The only way you could actually see how Fred and Barney got called a match is for one of them to run ADSA on his/her kit and email you the result. The other take-away from this is that there may be more people who share the same ancestor with the three of you (you, Fred, and Barney). Little Lulu might match Fred and Barney on some segment (or portion of a segment) that you don't have, and it came from the same ancestor as one or both of the segments you share with Fred and Barney. You would never know it. So the matches of your matches might be useful to know. Unfortunately though, with ICW all you get are the matches of your matches that also happen to match you. That's a subset of the larger set.
The procedure I follow when analyzing my chromosomes is to start with the longest segments I can find and work my way down to the smaller ones. The longer ones are more likely to have a relationship with me that is closer and therefore easier to find in my paper tree and the trees of my matches. Look for multiple overlaping segments that all ICW with each other (as described in the example above), then hover over the names of the people associated with them on the far left and see whether there are any ancestral surnames among them that you recognize. You might also look for ICW names that are not in the group by hovering over the segments. The segment pop-up shows other chromosomes where you match the same people - you can then find the table for those chromosomes and see if any of the same names show up there. Also, finding the same email address for different match names is sometimes a good indicator that those persons are all pretty close relatives (a daughter testing herself and her parents and siblings, for example).
Once you've generated a chart, if you want to go back to the input form and change any of the parameters, you'll find that using the browser's BACK button will preserve the inputs you already had entered, allowing you to change just the ones you want to change and re-run the tool. If you reload the page or navigate to its web address again, everything will return to the default settings and you will have to also respecify your files or kit number.
Hopefully this explanation is helpful. I have several ideas for enhancements to this, including a sort of "natural language" input that will allow more flexibility in setting initial parameters and adding more (eg. bolding or coloring the text for segments over a certain length or after a given match date, or hiding matches with more than 2000 total shared cMs (close relatives), etc.) I may also create different sort orders and ways of slicing and dicing the Match file or printing out the entire ICW table. I was also considering allowing you to enter your own comments or annotations to the data and store it on your computer in a 4th csv file or on the server. We are also planning to support 23andMe (and eventually Ancestry) files at some point. I imagine there are quite a few people out there who don't know how to use a spreadsheet, and maybe this tool can be helpful to them as an alternative to learning Excel. I guess my enthusiasm for future enhancements will depend on how enthusiastic you all are about using it!
If you have any suggestions for improvements or for goodness sake, if you find any bugs, please contact me. Many people have used ADSA successfully, but you may run into a problem I haven't seen yet. If you find something like that, please let me know right away so I can improve the program.
Don Worth
Oxnard, California
worth@ucla.edu
Read More About ADSA
- "Introducing the Autosomal DNA Segment Analyzer", DNA-eXplained, Roberta Estes, 9 Jan 2014
- "Autosomal DNA Segment Analyzer (ADSA) – No Spreadsheets Required! (PART 1)", Genealogy Junkie, Sue Griffith, 15 Jan 2014
- "The Worthwhile Autosomal DNA Segment Analyzer", Family History Research by Jody, Jody Lutter, 15 Jan 2014
READ THIS IF THINGS DON'T WORK FOR YOU We'll start with an explanation of the error messages that are possible: Invalid Chromosome - must be 1-22, X, or blank Base pair range can only be used with a single chromosome One or both of the base pair values is not numeric Ending base pair value is before starting base pair value Non-numeric or negative segment cMs Minimum Non-numeric or negative SNPS Minimum Non-numeric or negative most distant cousin Non-numeric or invalid graph width in pixels Chromosome Browser file name not specified or could not be read Matches file name not specified or could not be read File names do not appear to start with a kit number CB and Matches files do not start with the same kit number - they may not be proper DNAgedcom files ICW file could not be read ICW file does not start with the kit number - it may not be a proper DNAgedcom file Kit not found on server Matches file missing from kit directory on server Chromosome Browser file missing from kit directory on server ICW file missing from kit directory on server Missing header row or incorrect format for Matches file Empty or invalid Matches file Missing header row or incorrect format for Chromosome Browser file WARNING: "somenameorother" in Chromosome Browser file not found in Matches file. Match data unavailable. There are no matching segments that fit the criterea to graph WARNING: "somenameorother" or "someothernameorother" in ICW file was not found in Matches file. Skipping this ICW row. TROUBLESHOOTING TIPS The following are a few things that have been reported to support@DNAgedcom.com. Your problem may be among them. When downloading files from DNAgedcom.com... If your DNAgedcom download doesn't work correctly, first make sure you are entering the FamilyTreeDNA kit number and password on the download panel. Do not use the DNAgedcom user ID and password you created to logon to DNAgedcom. You may be prompted with them, but you need to replace them with the FamilyTreeDNA information instead. If the download seems to take too long (allow 15 minutes at least - more if you have Ashkenazi ancestry) or hangs, try looking for your files on DNAgedcom's server. Before it starts downloading your files it stores the complete files on its server. You can find them by hovering your cursor over the MEMBERS tab (top right on the home screen) and selecting VIEW FILES. You will see a list of folders on the left - click on the one with your kit number to open it. You should see your three files in the panel on the right. Double-click on each file in turn to download it to your computer. Make sure all three files are there (and not in a folder named "building") and that they have lengths that are not zero. If they are not in the kit folder or one or more is missing or zero length, then the download did not complete. Try downloading using DNAgedcom to download from Family Tree DNA again. One thing we are noticing is that a lot of people are impatient. If the download times out, go to VIEW FILES and look in the folders there. Give it some time to finish downloading the old set of files before starting the download over again. It's also possible that some people might be getting intermittent timeouts on the internet and that causes the connection to drop. The program may continue to download in the background. Again, give it a little bit of time. Finally, DNAGedcom.com is at the mercy of the backend system (in this case Family Tree DNA) when downloading data. Please give it time to run. If it doesn’t work the first time, often you can come back the next day or late at night or early in the morning and it will work fine. There are a LOT of calls to the back end web site to get this data, so being patient is the key. If you still can't get the downloads to work, contact the nice people at DNAgedcom.com and they will assist you. Here is their email address: support@dnagedcom.com After you click the GRAPH button on the ADSA input screen... If you get an error message like "Matches file name not specified or could not be read" you probably forgot to tell ADSA where to find that file so it could be read. Check to make sure all of the three files have been located with the BROWSE buttons at the bottom of the panel (omitting the ICW file if you wish to). ADSA will also check the file names to make sure they all start with the same kit number and will look for the header row in the Matches and Chromosome Browser files just to make sure it's not reading your daughter's term paper. If you don't see prompts for the three files, and you see a prompt for kit number instead, that means you are logged onto DNAgedcom. That's a good thing! Enter your kit number and probably your files are already on the server and ADSA will read them (more quickly) from there. If you click on GRAPH and, after a while, you get a blank screen or a 500 Server Internal Error or something of the kind, it means the server timed out trying to upload or read and process your files. One or more of your files may be too large for the server to handle. In some cases (such as if you have Ashkenazi ancestry) you may have far too many segments and ICWs for ADSA to handle gracefully. If you have not downloaded your files with DNAgedcom recently, it is recommended that you re-download them now. Updates to DNAgedcom made around 1/13/2014 have resulted in a much smaller ICW file than formerly. It is also recommended that you use ADSA by logging on to DNAgedcom first and accessing your files on the server with your kit number. That way they do not have to be reuploaded to the server before ADSA can begin processing them. Using the most distant cousin input value is also a way to make ADSA run faster with less possibility of a timeout. But for some really, really large Ashkenazi files, ADSA just may not work as it is now designed. You may have to find a way to manully subset your files in some fashion before feeding them into ADSA. Note that it is possible to upload files to your account's directory on the server using the VIEW FILES panel in the MEMBERS menu after you have edited them. If you see WARNINGS... A brief word about troubleshooting data problems. In the process of debugging the program I found some inconsistencies between the three files for Match Name (aka Full Name) and ICW Name, etc. For the tool to work, it is pretty dependent on being able to relate Chromosome Browser rows and ICW rows to Match file rows. The only way to do that is to compare the names it finds in the ICW and Chromosome Browser files to those in the Match file. Sometimes (rarely) the names get spelled differently in different files. The tool attempts to clean out any punctuation and extra spaces in the names before comparing them and that often makes things match, but I recently ran across a kit that had files that had some problems in them. So, if you get a lot of warnings, be aware that your tables may be missing some data for the people listed in the warnings because the tool couldn't find them in the Match file. I have tried to make the tool as robust as possible and to tolerate unexpected data. But there are some things I couldn't completely square away. For example, if more than one of your matches left their name blank, then all segments and ICWs with blank names will map to the first blank-named match in the Matches file. (For that reason I would suggest you ignore any segments with blank names.) Of course, you also have the option to open your files in a text editor and look for the source of problems, using the warnings and error messages to guide you, and manually correct them before running ADSA. By the way, if you decide to edit any of the three files on a Mac before loading them into ADSA, be sure to save them in Windows CSV format if you are on Mac OSX or Linux. If you save them as simply CSV they won't read in correctly because Windows has its own way of doing end of line characters. If none of this solves your problem, feel free to email me at worth@ucla.edu and we'll figure it out together. |